GPS faculty recently published a paper in Clinical Genetics that describes how NGS is being used in PIK3CA related overgrowth syndromes.
Utility of Clinical High-Depth Next Generation Sequencing for Somatic Variant Detection in the PIK3CA Related Overgrowth Spectrum. Hucthagowder V, Shenoy A, Corliss M, Vigh-Conrad KA, Storer C, Grange DK, Cottrell CE. Clin Genet. 2016 Jun 16. PubMedID 27307077
View all of our publications.
Abstract from PubMed
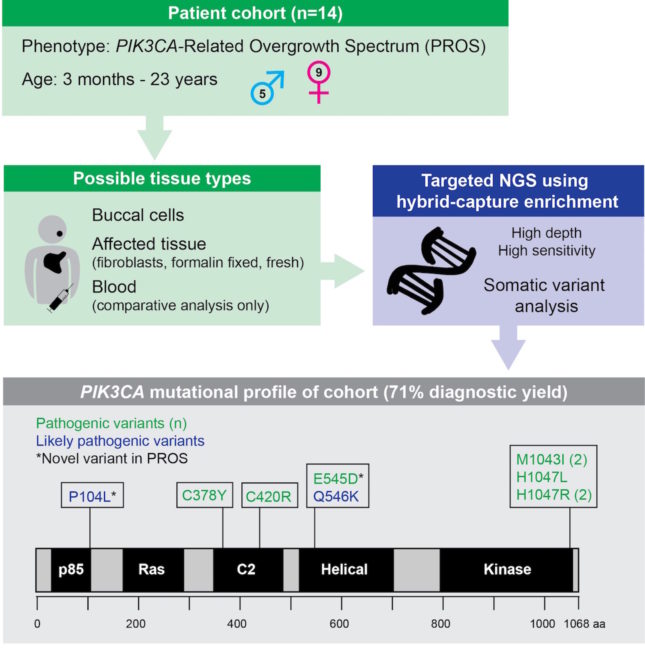
Next-generation sequencing (NGS) has revolutionized the approach of studying sequence variation, and has been well described in the clinical laboratory setting for the detection of constitutional alterations, as well as somatic tumor associated variants. It is increasingly recognized that post-zygotic somatic alteration can be associated with congenital phenotypic abnormalities. Variation within the PI3K/AKT/mTOR pathway, including PIK3CA, has been described in somatic overgrowth syndromes and vascular malformations. Detection of PIK3CA somatic alteration is challenging due to low variant allele frequency (VAF) along with the need to assay involved tissue, thus necessitating a highly-sensitive methodology. Here we describe the utility of target hybrid capture coupled with NGS for the identification of somatic variation in the PIK3CA-Related Overgrowth Spectrum (PROS) among 14 patients submitted for clinical testing. Assay detection of low allelic fraction variation is coverage dependent with >90% sensitivity at 400x unique read depth for VAF of 10%, and approaching 100% at 1000x. Average read depth among the patient dataset across PIK3CA coding regions was 788.4. The diagnostic yield among this cohort was 71%, including the detection of two PIK3CA alterations novel in the setting of PROS. This report expands the mutational scope and phenotypic attributes of PROS disorders.